Association of Oral Submucous Fibrosis Risk with GSTM1 and GSTT1 Gene Polymorphisms
By Abdul Mannan1, Munir Ahmad Bhinder1, Haleema Sadia2, Zainab Yousaf1, Zawar Hussain3, Muhammad Akram4, Madiha Shakoor1, Muhammad Yasir Zahoor5, Rahat Abdul Rehman6Affiliations
doi: 10.29271/jcpsp.2024.03.296ABSTRACT
Objective: To determine the association of GSTM1 and GSTT1 polymorphisms with oral submucous fibrosis (OSF).
Study Design: A case-control study.
Place and Duration of the Study: Department of Human Genetics and Molecular Biology, University of Health Sciences, Lahore and Oral and Maxillofacial Surgery Department, de Montmorency, College of Dentistry/ Punjab Dental Hospital, Lahore, Pakistan, from 1st April 2019 to 31st April 2020.
Methodology: OSF patients were diagnosed with different clinical staging of mouth opening by Vernier caliper with the help of a professional dentist in the Department of Oral and Maxillofacial, de Montmorency, College of Dentistry, Lahore. One hundred and eight blood samples of OSF patients and 108 samples of normal controls were collected. Genomic DNA was obtained from whole-blood extraction. Multiplex PCR amplification using GSTM1, GSTT1, and β -Globin gene primers was performed.
Results: GSTM1 and GSTT1 null genotypes frequencies were found in 43.5% (47/108) and 13.9% (15/108) of controls, whereas 54.6% (59/108) and 25.9% (28/108) of OSF patients, respectively. OSF patients had a greater frequency rate of GSTM1 and GSTT1 null genotypes than controls [OR 1.56, 95% CI 0.91–2.67 (p=0.13)] and [OR 2.17, 95% CI 1.08–4.34 (p=0.04)], respectively. The GSTT1 genotype was found statistically significant with OSF (p=0.05), and risk was also determined. The cumulative effect of null genotypes of GSTM1/GSTT1 did not show any association with the controls and in OSF patients. Proportions of active and null alleles of the patient group were; 86.1%/13.9%; and in control, it was 92.6%/7.4% (OR = 2.01; CI: 0.82–4.97; p=0.18), respectively.
Conclusion: The study determined a statistically significant association of GSTT1 gene polymorphism with OSF.
Key Words: Oral submucous fibrosis, GSTM1, GSTT1, Gene polymorphisms, Genetic risk.
INTRODUCTION
Oral Submucous Fibrosis (OSF) is a chronic, complicated, and precancerous disease of the mouth cavity caused by the chewing habit of areca nuts. Collagen begins in the lamina propria and prolongs to the submucosa, muscle, and beyond in this condition, which is linked to a juxta-epithelial inflammatory reaction in the mouth cavity, which restricts mouth opening and difficulty in swallowing.1 The OSF has been found most commonly in the mouth cavity's buccal mucosa and retromolar region, followed by the soft palate, faucial pillars, floor of the mouth, tongue, labial mucosa, and gingiva.2
According to the World Health Organisation (WHO), five million people survive through OSF worldwide.3 OSF in any population exists differently by ethnicity and region. Diet, habits, and culture majorly affect such diseases.1 Due to the migration of endemic betel quid chewers, OSF has become a public health issue in many parts of the world, such as the United Kingdom, South Africa, and numerous Southeast Asian countries.4 The highest prevalence of OSF has been seen in South and Southeast Asia populations.5 The exact cause of OSF is unclear, but the disease is complex and multifactorial. OSF tends to be caused by various reasons, including excessive chili intake, dietary deficiencies, genetic predispositions, and auto-immune diseases.6,7 One of OSF's most significant risk factors is chewing a betel quid containing areca nuts. OSF production is connected to betel quid's number of areca nuts and the extent and time of chewing.8
Glutathione S-transferases (GSTs) are involved in metabolising chemical carcinogens, such as those found in cigarette smoke and areca nuts. By deactivating or detoxifying electrophilic carcinogens, GSTs prevent the carcinogenic process from starting. Certain kinds of GSTs are expressed and started in precancerous and cancerous cells throughout the initiation and promotion stages.9 Glutathione S-transferases mu1 (GSTM1) and Glutathione S-transferases theta 1 (GSTT1) are polymorphic, and their deleted variants (null genotypes) result in complete functional loss.
The GST gene family is a dimeric enzyme structure that has been found in a variety of tissues. GST-class enzymes conjugate various reactivate electrophiles using a reduced glutathione (GSH) stage.10,11 In addition, lowering their levels plays a vital role in cellular defense. GSTM1 and GSTT1 homozygous deletions result in the loss of GSTM1 and GSTT1 protein products.10 The objective of this study was to determine the association of GSTM1 and GSTT1 polymorphisms with OSF.
METHODOLOGY
In the present case-control study, 108 individuals with OSF and 108 individuals in the normal control group were recruited. These samples were genotyped for GSTM1 and GSTT1 gene polymorphisms. Informed consent was taken from the recruited individuals, and the study was approved by the Ethical Review Committee for Medical and Biomedical Research, University of Health Sciences, Lahore (UHS/REG-19/ERC/1862). Patients were registered in the Oral and Maxillofacial Surgery Department, de Montmorency, College of Dentistry/ Punjab Dental Hospital, Lahore, Pakistan. All patients were diagnosed with OSF.
Patients with OSF were diagnosed by a professional Dentist, both male and female patients with limited mouth opening with clinically established OSF of all ages were selected for the patient group while healthy oral mucosa from different pan shops with chewing habits, both males and females were included in the control group. Patients with immunocompromised conditions and patients with clinically diagnosed indurated ulcers in the oral cavity were excluded.
Genomic DNA extraction was done through the phenol-chloroform method, known as organic, on 5 ml of frozen blood samples. Multiplex PCR method was applied for the genotyping of GSTM1 and GSTT1 genes. The sequence of primers for GSTM1 was (Forward: 5’-GAACTCCCTGAAAAGCTAAGC-3 and Reverse: 5’-GTTGGGCTCAAATATACGGTGG-3), for GSTT1 gene was (Forward: 5’-TTCCTTACTGGTCCTCCATCTC-3' and Reverse 5’-TCACCGGATCATGGAAACCA-3'), and for β-Globin gene was (Forward:5’-CAACTTCATCCACGTTCACC-3 and Reverse: 5'- GAAGAGCCAAGGACAGGTAC-3). The base products for GSTM1, GSTT1, and β-Globin genes were 215 base pairs (Bp), 480 Bp, and 268 Bp, respectively.
Gene polymorphisms of GSTM1 and GSTT1 sequences were identified by the sequences derived from the Ensemble Genome browser. Primers for amplification were synthesised commercially. The forward and reverse primers of gene polymorphisms of GSTM1, GSTT1, and β-globin genes were preferentially amplified in a multiplex manner; each sample was found to have β-globin gene amplification. The other two genes GSTM1 and GSTT1 showed null or active genotypes. The purpose of β-globin gene amplification is whether the PCR reaction was successful or not.
The total reaction volume of 17 µl for each sample reaction mixture contained 2 µl genomic DNA, 0.5 µl of each forward and reverse primer for each gene GSTM1, GSTT1, and β-Globin, respectively, 5 µl of master mix (10X PCR buffer, 15 mM MgCl2, 20mM of dNTP mix and 0.5 units of Taq DNA Polymerase), and 7 µl of dH20. Denaturation of DNA was initiated at 95°C for 3 minutes. An annealing temperature of 58°C was optimised for each of the six primers with a time of 30 seconds. The extension was carried at 72°C for 01 minutes, and the cycle was repeated. Every PCR reaction has about 35 cycles, and each run is initiated with a starting hold of 45 seconds at 95°C. A 2% agarose gel containing ethidium bromide was used to examine the amplified results (Figure 1).
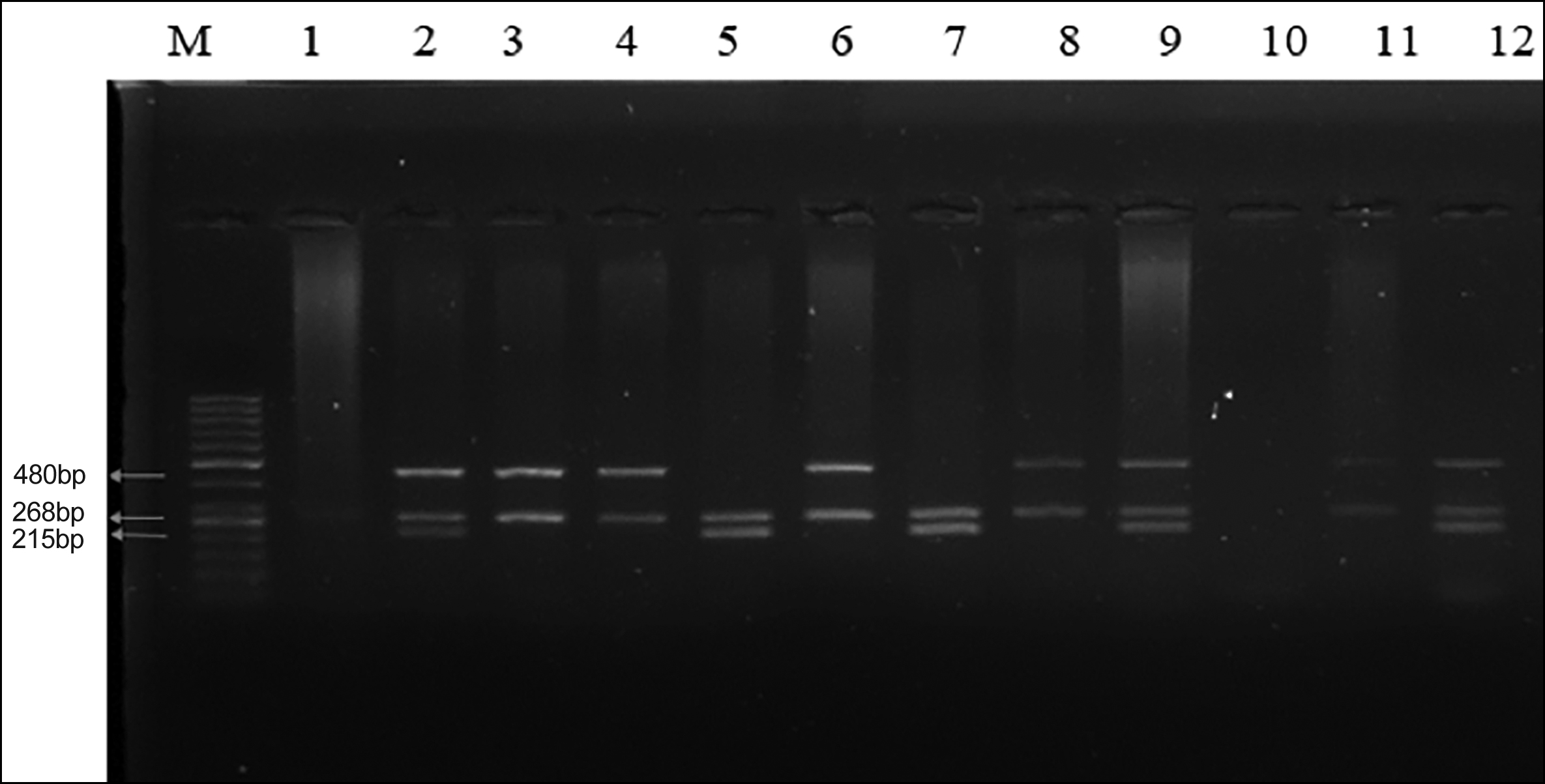 Figure 1: GSTT1 null genotype is shown by the absence of a 480-bp band; the absence of a 215-bp band indicates GSTM1 null genotype; β-Globin was coamplified in all samples. M marker represents the 50 bp DNA Ladder. Lanes 2, 9, and 12; represent GSTM1 and GSTT1 positive genotypes. Lanes 3, 4, 6, 8, and 11; represent GSTM1 null genotype, and lane 10; represents negative control. Lane 1, GSTM1 and GSTT1 null genotype.
Figure 1: GSTT1 null genotype is shown by the absence of a 480-bp band; the absence of a 215-bp band indicates GSTM1 null genotype; β-Globin was coamplified in all samples. M marker represents the 50 bp DNA Ladder. Lanes 2, 9, and 12; represent GSTM1 and GSTT1 positive genotypes. Lanes 3, 4, 6, 8, and 11; represent GSTM1 null genotype, and lane 10; represents negative control. Lane 1, GSTM1 and GSTT1 null genotype.
The data were entered and analysed by using Statistical Package for Social Sciences (SPSS) version 26.0. Mean + SD and median with interquartile range were provided for quantitative variables. In both OSF and control groups, the polymorphism was confirmed by using Hardy-Weinberg expectations. To compare the two groups, the chi-square test and the t-test were used. The categorical variables were expressed as counts and percentages. Using univariate analysis, the association between the GSTM1 and GSTT1 variant genotypes and the risk of OSF is evaluated using odds ratios (ORs) with 95% confidence intervals (CIs). A p-value of less than and equal to 0.05 was considered statistically significant.
RESULTS
The frequency of GSTM1 and GSTT1 null genotypes was higher in OSF patients (OR=1.56, p=0.13) than in the control group (OR=2.17, p=0.04). The cumulative effect of combined GSTM1/GSTT1 genotypes was also estimated using OR with varied combinations of the two polymorphisms. The frequency of dual null genotypes was not significantly measured with the double null genotype. The risk of OSF association with the GSTM1, GSTT1, and GSTM1/GSTT1 dual genotypes is shown in Table I.
Table I: Risk of OSF association with the GSTM1, GSTT1, and GSTM1/GSTT1 dual genotypes.
|
Genotypes |
OSF (n, %) |
Controls (n, %) |
OR (95% CI) |
p-value (Chi-square) |
|
GSTM1 |
||||
|
Active (+/+, +/-) |
49 (45.4%) |
61 (56.5%) |
1 |
0.13 |
|
Null (-/-) |
59 (54.6%) |
47 (43.5%) |
1.56 (0.91-2.67) |
|
|
GSTT1 |
||||
|
Active (+/+, +/-) |
80 (74.1%) |
93 (86.1%) |
1 |
0.04* |
|
Null (-/-) |
28 (25.9%) |
15 (13.9%) |
2.17 (1.08-4.34) |
|
|
GSTM1+GSTT1 |
||||
|
Active (+/+, +/-) |
93 (86.1%) |
100 (92.6%) |
1 |
0.18 |
|
Null (-/-) |
15 (13.9%) |
8 (7.4%) |
2.01 (0.82-4.97) |
|
|
*A p-value of <0.05 was considered significant. |
||||
Table II: Demographic and clinical characteristics of OSF cases and control group.
|
Characteristics |
OSF patients (n = 108) |
Controls (n = 108) |
Chi-square |
|
Age |
25.89+8.69 |
28.80+10.54 |
0.04* |
|
Gender (n, %) |
|||
|
Males |
76 (70.4%) |
74 (68.5%) |
0.88 |
|
Females |
32 (29.6%) |
34 (31.5%) |
|
|
Socioeconomic status (n, %) |
|||
|
Lower class |
79 (73.1%) |
87 (80.6%) |
0.26 |
|
Middle class |
29 (26.9%) |
21 (19.4%) |
|
|
Cast |
|||
|
Arrain |
29 (26.9%) |
23 (21.3%) |
0.16 |
|
Jutt |
8 (7.5%) |
18 (16.7 %) |
|
|
Rajput |
25 (23.1%) |
20 (18.5 %) |
|
|
Others |
46 (42.6%) |
47 (43.5 %) |
|
|
Mouth opening (n, %) |
|||
|
1-10 mm |
10 (9.3%) |
- |
- |
|
11-20 mm |
73 (67.6%) |
- |
|
|
21-30 mm |
24 (22.1%) |
- |
|
|
Use of supari (n, %) |
|||
|
Yes |
98 (90.7%) |
- |
- |
|
No |
10 (9.3%) |
- |
|
|
Use of pan (n, %) |
|||
|
Yes |
54 (50%) |
- |
- |
|
No |
54 (50%) |
- |
|
|
Use of gutka (n, %) |
|||
|
Yes |
9 (8.3%) |
- |
- |
|
No |
99 (91.7%) |
- |
|
|
*A p-value of <0.05 was considered significant. |
|||
The dual GSTM1/GSTT1 genotypes proportions of active and null alleles of the patient group were 86.1%, and 13.9%, respectively, and in the control group, it was 92.6% and 7.4%, respectively (OR=2.01, p=0.18).
No significant association was seen in demographic features like gender (p=0.88), socioeconomic status (p=0.26), cast (p=0.16), and the mean age (p=0.049), in OSF patients and control group. Demographic and clinical characteristics of OSF patients and the control group are shown in Table II. Clinical characteristics and different life habits of OSF cases were analysed statistically, like a mouth opening and supari, pan, and gutka. The percentage of mouth opening with 1-10 mm was 9.3%, with 11-20 mm was 67.6%, and with 21-30 mm was 22.1%, respectively, the percentage of use of supari, pan, and gutka who had chewing habit of it was 90.7%, 50%, and 8.3%, respectively.
No significant association was found among active and null GSTM1 genotypes in demographic features like gender (p=0.40), socioeconomic status (p=0.470), cast (p=0.76), and mean age (p=0.06) in OSF patients. The mouth opening habit (p=0.18), use of supari (p=0.51), use of pan (p=0.24), and use of gutka (p=0.18), were also found statistically non-significant with active and null genotypes of GSTM1 gene.
No significant difference was found among active and null GSTT1 genotypes in demographic features like gender (p=0.99), socioeconomic status (p=0.09), cast (p=0.62), and mean age (p=0.14) in OSF patients. The mouth opening habit (p=0.22), use of supari (p=0.45), use of pan (p=0.27), and the use of gutka (p=0.69) were also found statistically non-significant with active and null genotypes of GSTT1 gene.
Table III: Association of GSTM1, GSTT1, and GSTM1-GSTT1 combined genotypes with demographic and clinical characteristics in OSF patients.
|
Characteristics |
GSTM1 polymorphism |
p-value (t-test) |
GSTT1 polymorphism |
p-value (t-test) |
GSTM1-GSTT1 polymorphism |
p-value (t-test) |
|||
|
Active (+/+, +/-) (n = 49) |
Null (-/-) (n = 59) |
Active (+/+, +/-) (n = 80) |
Null (-/-) (n = 28) |
Active (+/+, +/-) (n = 93) |
Null (-/-) (n = 15) |
||||
|
Age |
23.20+5.98 |
28.12+9.92 |
0.06 |
25.47+8.71 |
27.11+8.83 |
0.14 |
- |
- |
- |
|
Gender (n, %) |
|||||||||
|
Males |
32 (65.3%) |
44 (74.6%) |
0.40 |
56 (70%) |
20 (71.4%) |
>0.99 |
63 (67.7%) |
13 (86.7%) |
0.22 |
|
Females |
17 (34.7%) |
15 (25.4%) |
24 (30%) |
8 (28.6%) |
30 (32.3%) |
2 (13.3%) |
|||
|
Socioeconomic status (n, %) |
|||||||||
|
Lower class |
38 (77.6%) |
41 (69.5%) |
0.47 |
55 (68.8%) |
24 (85.7%) |
0.09 |
66 (71%) |
13 (86.7%) |
0.35 |
|
Middle class |
11 (22.4%) |
18 (30.5%) |
25 (31.2%) |
4 (14.3%) |
27 (29%) |
2 (13.3%) |
|||
|
Cast |
|||||||||
|
Arrain |
11 (22.4 %) |
18 (30.5 %) |
0.76 |
21 (26.3%) |
8 (28.6%) |
0.62 |
25 (26.9%) |
4 (26.7%) |
0.98 |
|
Jutt |
4 (8.2 %) |
4 (6.8 %) |
7 (8.8%) |
1 (3.6%) |
7 (7.5%) |
1 (6.7%) |
|||
|
Rajput |
11 (22.4%) |
14 (23.7 %) |
20 (25%) |
5 (17.9%) |
21 (22.6%) |
4 (26.7%) |
|||
|
Others |
23 (46.9 %) |
23 (39%) |
32 (40%) |
14 (50%) |
40 (43%) |
6 (40%) |
|||
|
Mouth opening (n, %) |
|||||||||
|
1-10 mm |
2 (4.2%) |
8 (13.6%) |
0.18 |
7 (8.9%) |
3 (10.7%) |
0.22 |
8 (8.7%) |
2 (13.3%) |
0.28 |
|
11-20 mm |
33 (68.8%) |
40 (67.8%) |
51 (64.5%) |
22 (78.6%) |
61 (66.3%) |
12 (80%) |
|||
|
21-30 mm |
13 (27%) |
11 (18.6%) |
21 (26.6%) |
3 (10.7%) |
23 (25%) |
1 (6.7%) |
|||
|
Use of supari (n, %) |
|||||||||
|
Yes |
43 (87.8%) |
55 (93.2%) |
0.51 |
71 (88.7%) |
27 (96.4%) |
0.45 |
83 (89.2%) |
15 (100%) |
0.35 |
|
No |
6 (12.2%) |
4 (6.8%) |
9 (11.3%) |
1 (3.6%) |
10 (10.8%) |
0 (0%) |
|||
|
Use of pan (n, %) |
|||||||||
|
Yes |
28 (57.1%) |
26 (44.1%) |
0.24 |
37 (46.3%) |
17 (60.7%) |
0.27 |
44 (47.3%) |
10 (66.7%) |
0.26 |
|
No |
21 (42.9%) |
33 (55.9%) |
43 (53.7%) |
11 (39.3%) |
49 (52.7%) |
5 (33.3%) |
|||
|
Use of gutka (n, %) |
|||||||||
|
Yes |
2 (4.1%) |
7 (11.9%) |
0.18 |
6 (7.5%) |
3 (10.7%) |
0.69 |
8 (8.6%) |
1 (6.7%) |
>0.99 |
|
No |
47 (95.9%) |
52 (88.1%) |
74 (92.5%) |
25 (89.3%) |
85 (91.4%) |
14 (93.3%) |
|||
|
*A p-value of <0.05 was considered significant. |
|||||||||
No significant difference was found among active and null GSTM1-GSTT1 combined genotypes in demographic features like gender (p=0.22), socioeconomic status (p=0.35), cast (p=0.98), and the mean age (p=0.27) in OSF patients. The mouth opening habit (p=0.28), use of supari (p=0.35), use of pan (p=0.26), and the use of gutka (p=0.99) were also found statistically non-significant with active and null genotypes of GSTM1-GSTT1 combined genotypes (Table III).
DISCUSSION
OSF is a chronic, complicated, precancerous (1% trans-formation risk) disease of the oral cavity linked to the mastication of betel quid-containing areca nuts.12 In this disease, collagen deposits in the lamina propria and spreads to the submucosa, muscle, and beyond, which is linked to a juxta epithelial inflammatory reaction in the mouth.13 The buccal mucosa and retromolar area are the most prevalent sites for OSF in the mouth cavity, followed by the soft palate, faucial pillars, floor of the mouth, tongue, labial mucosa, and gingiva.14 OSF is usually accompanied by a juxta-epithelial inflammatory response and fibroblastic changes in the lamina propria, the oral mucosa becomes stiff due to epithelial atrophy, producing trismus and trouble swallowing.15
OSF is a precursor to oral cancer; it has a high mortality rate, especially squamous cell carcinoma, which occurs in 7.6% of cases.16 Malignant transformation rates of 1.9-9% have been reported in several studies with shorter follow-up durations.17 People living with OSF are 19 times more likely to acquire oral squamous cell carcinoma than healthy people.18 A study showed that10 GSTM1 null genotype and antioxidants might be associated with the malignant transformation of the oral pre-cancers. The lack of the GSTT1 gene raises the incidence of oral lesions by fourfold because the frequency of the GSTT1 null genotype was shown to be substantially (p=<0.05) greater in cases than in controls.19
Another study demonstrated the association of the GSTM1 null allele with OSF. Due to varied patterns of association in different populations, such analysis is needed in the remaining people.20 OSF patients showed a greater incidence of GSTM1 and GSTT1 null genotypes (15/90) (16.6%).21 Individuals who had both the GSTM1 and GSTT1 null genotypes had a 7.5-fold greater risk of OSF (OR=7.5) than those who only had the GSTM1 null genotype.
The present study was conducted to generate (GSTM1, GSTT1) association data from Pakistani populations. The present study describes the association of the GSTT1 null allele with OSF in Pakistani patients. Although the GSTM1 null allele frequency is higher in the patient than in the control group, this difference is not statistically significant. The study emphasises the significance of this polymorphism and the necessity for more research into OSF development. This research may help to identify the biomarkers for the molecular diagnosis of OSF and the prediction of disease susceptibility, thus allowing for early therapeutic intervention in OSF patients of higher risk OSF patients in any community. Furthermore, this research might be expanded to see if GSTT1 and associated gene polymor-phisms individually influence OSF development.
CONCLUSION
The findings in this study show a link between the GSTT1 gene polymorphism and OSF susceptibility and favour the relationship of the GSTT1 genotype with OSF susceptibility. The GSTT1 gene polymorphisms are likely to contribute to the pathogenesis of OSF disease.
ETHICAL APPROVAL:
The study was approved by the Ethical Review Committee for Medical and Biomedical Research, University of Health Sciences, Lahore, Pakistan (UHS/REG-19/ERC/1862).
PATIENTS’ CONSENT:
The informed consent was obtained from all patients before the collection of data. The data were kept confidential and used only for study purposes.
COMPETING INTEREST:
The authors declared no conflict of interest.
AUTHORS’ CONTRIBUTION:
AM: Experimentation, data analysis, manuscript prepara-tion, and revision.
MAB: Concept, experimentation, data analysis, manuscript preparation and revision.
ZY: Data analysis, manuscript preparation, and revision.
HS, ZH, MA, MS, MYZ, RAR: Data analysis and manuscript preparation and revision.
All authors approved the final version of the manuscript to be published.
REFERENCES
- Shen YW, Shih YH, Fuh LJ, Shieh TM. Oral submucous fibrosis: A review on biomarkers, pathogenic mechanisms, and treatments. Int J Mol Sci 2020; 21(19):7231. doi: 10. 3390/ijms21197231.
- Gupta S, Hamid R, Ramasamy P, Ghosh S. Oral sub-mucous fibrosis-A review. Ann Romanian Soc Cell Biol 2022; 26(01):2875-85. Available from: https://www. annalsofrscb.ro/index.php/journal/article/view/11272.
- Gottipamula S, Sundarrajan S, Moorthy A, Padmanabhan S, N Sridhar K. Buccal mucosal epithelial cells down-regulate CTGF expression in buccal submucosal fibrosis fibro-blasts. J Maxillofac Oral Surg 2018; 17(2): 254-9. doi: 10.1007/s12663-017-1056-1.
- Wang M, Xiao C, Ni P, Yu JJ, Wang XW, Sun H. Correlation of betel quid with oral cancer from 1998 to 2017: A study based on bibliometric analysis. Chin Med J (Engl) 2018; 131(16):1975-82. doi: 10.4103/0366-6999.238140.
- Arora S, Squier C. Areca nut trade, globalisation and its health impact: Perspectives from India and South-east Asia. Perspect Public Health 2019; 139(1):44-8. doi: 10.1177/1757913918785398.
- Yadahalli R, Sarode GS, Sarode SC, Khan ZA, Vyas N, Kharat AH, et al. CC group of chemokines and associated gene expression of transcription factors: Deciphering immuno-pathogenetic aspect of oral submucous fibrosis. Dis Mon 2023; 69(1):101351. doi: 10.1016/j.disamonth. 2022.101351.
- Katira P, Panwar S, Rao D. Oral submucous fibrosis in pediatric patient: A case report with review of literature. Guident 2020; 14(1):38-41.
- Cirillo N, Duong PH, Er WT, Do CTN, De Silva MEH, Dong Y, et al. Are there betel quid mixtures less harmful than others? A scoping review of the association between different betel quid ingredients and the risk of oral submucous fibrosis. Biomolecules 2022; 12(5):664. doi: 10.3390/biom12050664.
- Ray JG, Chatterjee R, Chaudhuri K. Oral submucous fibrosis: A global challenge. Rising incidence, risk factors, management, and research priorities. Periodontol 2000 2019; 80(1):200-12. doi: 10.1111/prd.12277.
- Madhulatha G, Das S, Venkateswarlu N, Pujar A, Jyothy A, Munshi A. GSTM1 and GSTT1 null polymorphism and antioxidant levels in oral submucous fibrosis, leukoplakia and oral cancer patients among a South Indian population. J Oral Maxillofac Surg Med Pathol 2018; 30(2):169-74. doi:10.1016/j.ajoms.2017.11.008.
- Rai A, Siddiqui M, Parveen S, Parveen S, Rasheed A, Ali S. Molecular pathogenesis of oral submucous fibrosis: A critical appraisal. Biomed Pharmacol J 2019; 12(04): 2027-36. doi: doi:10.13005/bpj/1835.
- Campos MC, Tubau C, Segura S, González-Farré M, Iglesias-Sancho M, Fernández-Figueras MT, et al. Oral submucous fibrosis presenting with histopathological lichenoid changes as predominant feature: Report of five cases and review of the literature. J Cutan Pathol 2021; 48(11): 1392-6. doi: 10.1111/cup.14084.
- Angadi PV, Rao SS. Areca nut in pathogenesis of oral submucous fibrosis: Revisited. Oral Maxillofac Surg 2011; 15(1):1-9. doi: 10.1007/s10006-010-0219-8.
- Fry R, Goyal S, Pandher P, Chawla J. An approach to mana-gement of oral submucous fibrosis: Current status and review of literature. Int J Curr Res 2014; 6:10598-604. Available from: https://journalcra.com/sites/default/files/issue- pdf/7001.pdf
- Rajendran R, Rajeesh MP, Shaikh S, Shanthi, Pillai MR. Expression of matrix metalloproteinases (MMP-1, MMP-2 and MMP-9) and their inhibitors (TIMP-1 and TIMP-2) in oral submucous fibrosis. Indian J Dent Res 2006; 17(4): 161-6. doi: 10.4103/0970-9290.29870.
- Saleem Z, Shaikh AH, Zaman U, Ahmed S, Majeed MM, Kazmi A, et al. Estimation of salivary matrix metallo-proteinases-12 (MMP- 12) levels among patients presenting with oral submucous fibrosis and oral squamous cell carcinoma. BMC Oral Health 2021; 21(1):205. doi: 10. 1186/s12903-021-01571-7.
- Arakeri G, Patil SG, Aljabab AS, Lin KC, Merkx MAW, Gao S, Brennan PA. Oral submucous fibrosis: An update on pathophysiology of malignant transformation. J Oral Pathol Med 2017; 46(6):413-7. doi: 10.1111/jop.12582.
- Nethan ST, Gupta S, Warnakulasuriya S. Risk factors for oral squamous cell carcinoma in the Indian population. Microbes and oral squamous cell carcinoma: Springer; 2022. p. 9-40. doi:10.1007/978-981-19-0592-6_2.
- Tripathi P, Yadav SK, Sanyal S, Singh A. Polymorphism of two genes and oral lesion risk in North Indian population. Int J Cancer Res 2017; 13:84-8. doi: 10.3923/ijcr.2017. 84.88
- Ramachandran S, Balan A, Balaram P. Genetic poly-morphism of GSTM1 as risk factor for oral potentially malignant disorder. J Dent Med Sci 2017; 16(8):47-50 doi:10. 9790/0853-1608124750.
- Agrawal D, Gupta S, Agarwal D, Gupta OP, Agarwal M. Role of GSTM1 and GSTT1 polymorphism: Susceptibility to oral submucous fibrosis in the North Indian population. Oncology 2010; 79(3-4):181-6. doi: 10.1159/000318533.